This vignette shows how to smooth and thin WATLAS data.
# Packages
library(tools4watlas)
library(ggplot2)
# Path to csv with filtered data
data_path <- system.file(
"extdata", "watlas_data_filtered.csv",
package = "tools4watlas"
)
# Load data
data <- fread(data_path, yaml = TRUE)Median smooth data
To reduce error in the position data, a basic smoother such as a
median filter can be applied. the function
atl_median_smooth calculates the median coordinates within
a window of positions set by moving window.
# Smooth the data
data <- atl_median_smooth(data, moving_window = 5)The resulting table overwrites the smoothed coordinates in the
columns x and y and keeps the original ones in
the columns x_raw and y_raw.
Calculate speed and turning angle
After median filtering the data, the speeds need to be recalculated. We will also calculate turning angles.
Note: the distance between median smoothed positions can be 0 and therefore will produce NAs and a warning
# Recalculate speed
data <- atl_get_speed(data, type = c("in", "out"))Look at the data
This plot just shows one example of a raw and median smooted track.
# subset first tag
data_subset <- data[tag == data[1]$tag]
# subset some data to look at
from <- min(data_subset[, datetime]) + 1 * 3600
to <- min(data_subset[, datetime]) + 12 * 3600
data_subset <- data_subset[datetime %between% c(from, to)]
# Create basemap
bm <- atl_create_bm(data_subset)
# Plot
bm +
geom_path(
data = data_subset, aes(x_raw, y_raw),
color = "firebrick3", linewidth = 0.5
) +
geom_path(
data = data_subset, aes(x, y),
color = "black", linewidth = 0.5
) +
geom_point(
data = data_subset, aes(x_raw, y_raw),
color = "firebrick3", size = 1.2
) +
geom_point(
data = data_subset, aes(x, y),
color = "black", size = 1
)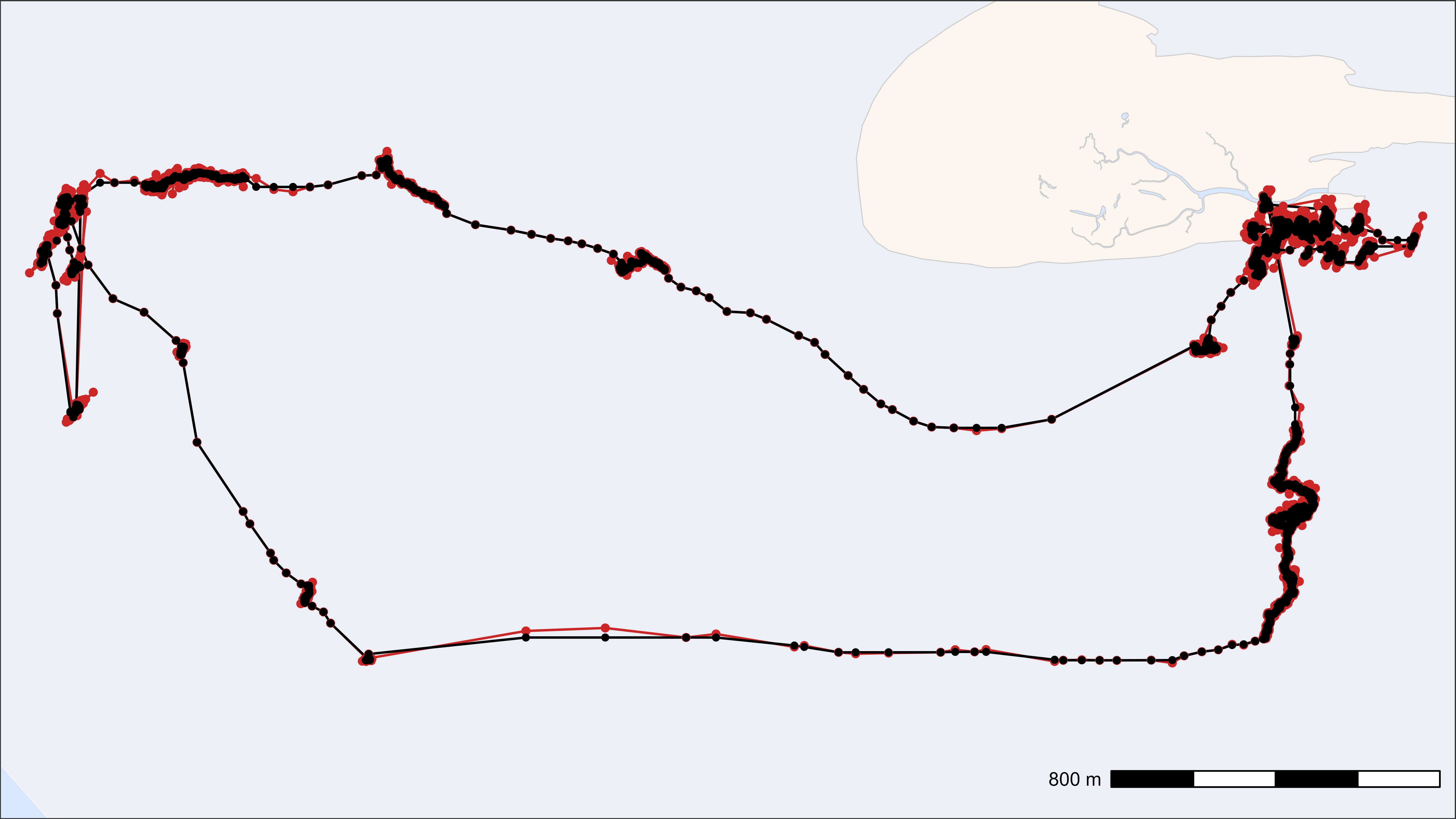
Smoothed track (black) on top of raw track (red)
Save data for the next steps
# Save data
fwrite(
data,
file = "../inst/extdata/watlas_data_smoothed.csv", yaml = TRUE
)Thin data
Depending on the desired analysis, it might make sense to thin data, either by aggregation or by subsampling. Both methods return fixed time steps (depending on the interval).
By aggregation
Returns the mean of all columns for each time step. The additional
column n_aggregated shows how many positions were
aggregated for this position. Time and datetime are returned rounded
down to the desired interval.
# Thin the data by aggregation with a 60-second interval
thinned_aggregated <- atl_thin_data(
data = data,
interval = 60,
id_columns = c("tag", "species"),
method = "aggregate"
)
# Show head of selected data
head(thinned_aggregated[, .(tag, time, datetime, x, y, n_aggregated)]) |>
knitr::kable(digits = 2)| tag | time | datetime | x | y | n_aggregated |
|---|---|---|---|---|---|
| 3027 | 1695438780 | 2023-09-23 03:13:00 | 650705.6 | 5902556 | 3 |
| 3027 | 1695439140 | 2023-09-23 03:19:00 | 650722.1 | 5902562 | 4 |
| 3027 | 1695439200 | 2023-09-23 03:20:00 | 650712.0 | 5902563 | 10 |
| 3027 | 1695439260 | 2023-09-23 03:21:00 | 650702.9 | 5902562 | 1 |
| 3027 | 1695439440 | 2023-09-23 03:24:00 | 650705.2 | 5902576 | 6 |
| 3027 | 1695439500 | 2023-09-23 03:25:00 | 650700.1 | 5902562 | 17 |
By subsampling
Returns the first position for each time step. The column
n_subsampled shows from how many positions this position
was sampled.
# Thin the data by subsampling with a 60-second interval
thinned_subsampled <- atl_thin_data(
data = data,
interval = 60,
id_columns = c("tag", "species"),
method = "subsample"
)
# Show head of selected data
head(thinned_subsampled[, .(tag, time, datetime, x, y, n_subsampled)]) |>
knitr::kable(digits = 2)| tag | time | datetime | x | y | n_subsampled |
|---|---|---|---|---|---|
| 3027 | 1695438802 | 2023-09-23 03:13:22 | 650705.6 | 5902556 | 3 |
| 3027 | 1695439189 | 2023-09-23 03:19:49 | 650721.0 | 5902559 | 4 |
| 3027 | 1695439201 | 2023-09-23 03:20:01 | 650723.1 | 5902564 | 10 |
| 3027 | 1695439261 | 2023-09-23 03:21:01 | 650702.9 | 5902562 | 1 |
| 3027 | 1695439477 | 2023-09-23 03:24:37 | 650702.8 | 5902562 | 6 |
| 3027 | 1695439501 | 2023-09-23 03:25:01 | 650709.9 | 5902598 | 17 |
