This article shows different ways on how to plot WATLAS data. In this example we use a simple basemap with the extend of the data, but any other type could be used too (see article: Create a basemap).
It first presents some examples of how to plot data grouped by tag ID, then by species and then how to make single plots with different options to quickly check the data, and how to plot heatmaps of the data. These examples can be customized as desired and should jsut give a quick starting point.
Plot grouped by tag ID
Simply colour points and the path with tag ID. Add for example the
number of tags as legend name if desired. When one needs individual
consistent colours between plots, one can use
atl_tag_cols(data$tag) to assign them (see second
example).
# load example data
data <- data_example
# create basemap
bm <- atl_create_bm(data, buffer = 800)
# plot points and tracks with standard ggplot colours
bm +
geom_path(
data = data, aes(x, y, colour = tag),
linewidth = 0.5, alpha = 0.1, show.legend = TRUE
) +
geom_point(
data = data, aes(x, y, colour = tag),
size = 0.5, alpha = 1, show.legend = TRUE
) +
scale_color_discrete(name = paste("N = ", length(unique(data$tag)))) +
theme(legend.position = "top")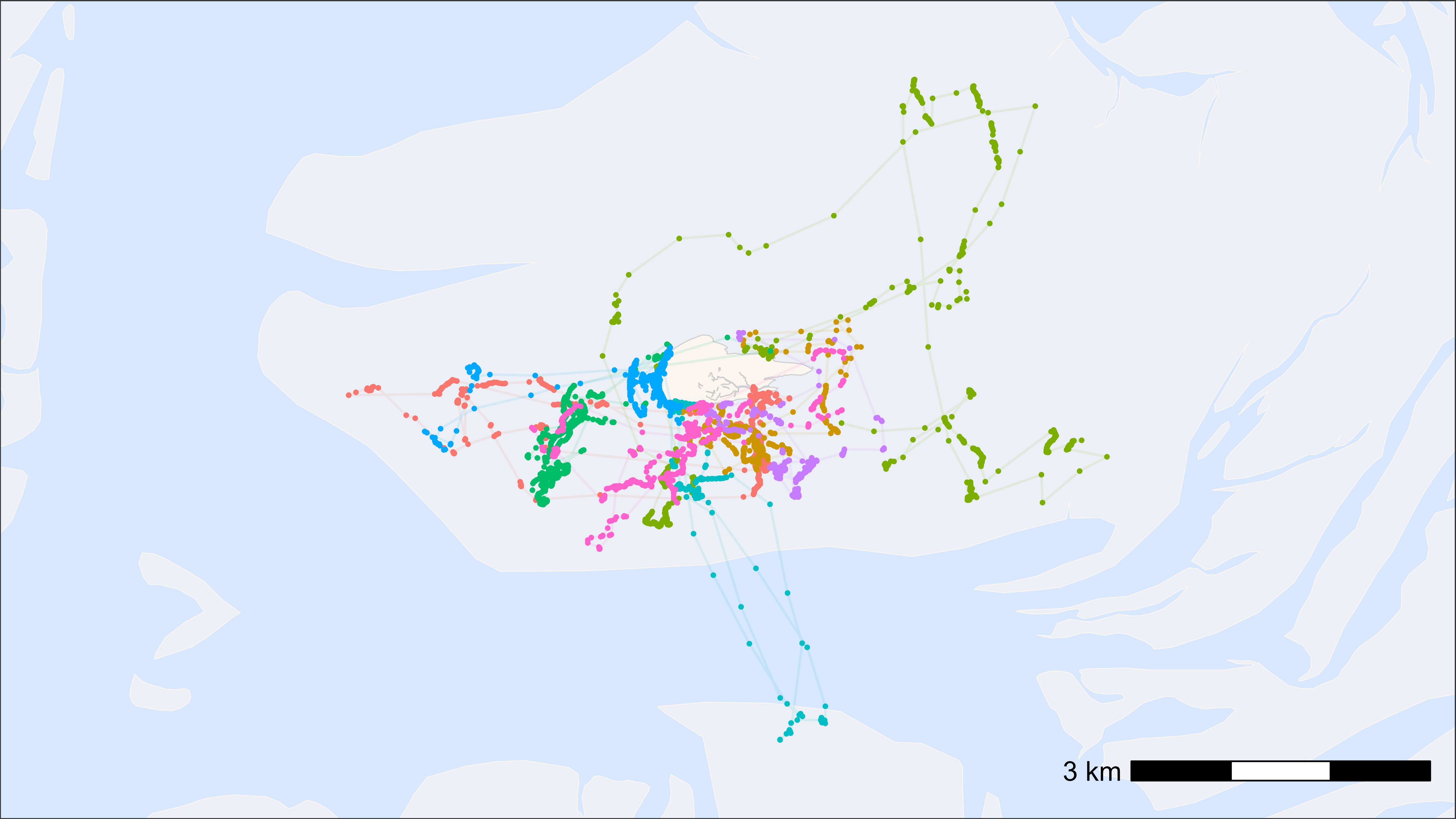
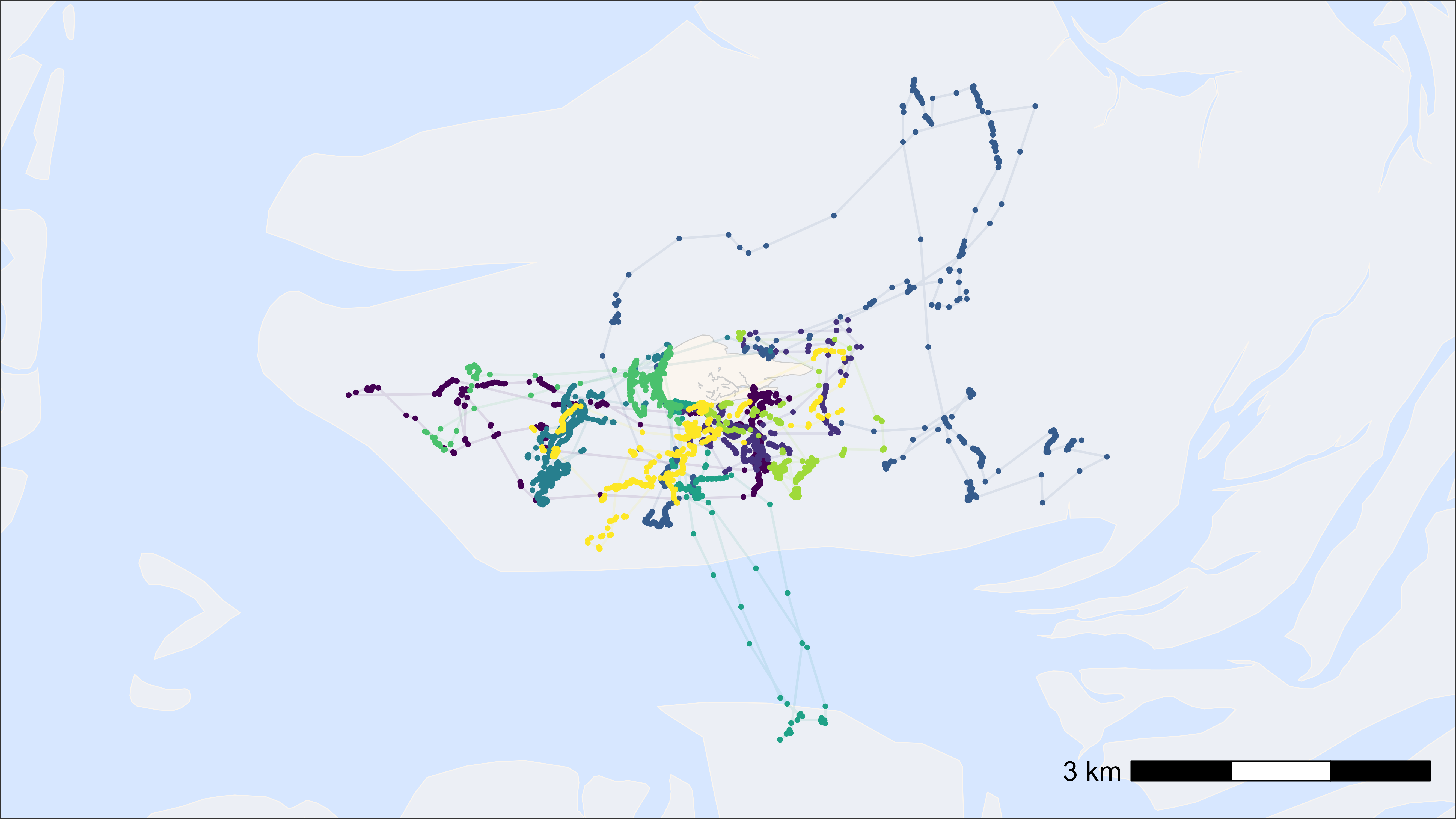
# plot points and tracks with fixed assigned colours (e.g. in animations)
bm +
geom_path(
data = data, aes(x, y, colour = tag),
linewidth = 0.5, alpha = 0.1, show.legend = FALSE
) +
geom_point(
data = data, aes(x, y, colour = tag),
size = 0.5, alpha = 1, show.legend = FALSE
) +
scale_color_manual(
values = atl_tag_cols(data$tag)
)
# plot points and tracks with viridis colour scale
bm +
geom_path(
data = data, aes(x, y, colour = tag),
linewidth = 0.5, alpha = 0.1, show.legend = FALSE
) +
geom_point(
data = data, aes(x, y, colour = tag),
size = 0.5, alpha = 1, show.legend = FALSE
) +
scale_color_viridis(discrete = TRUE)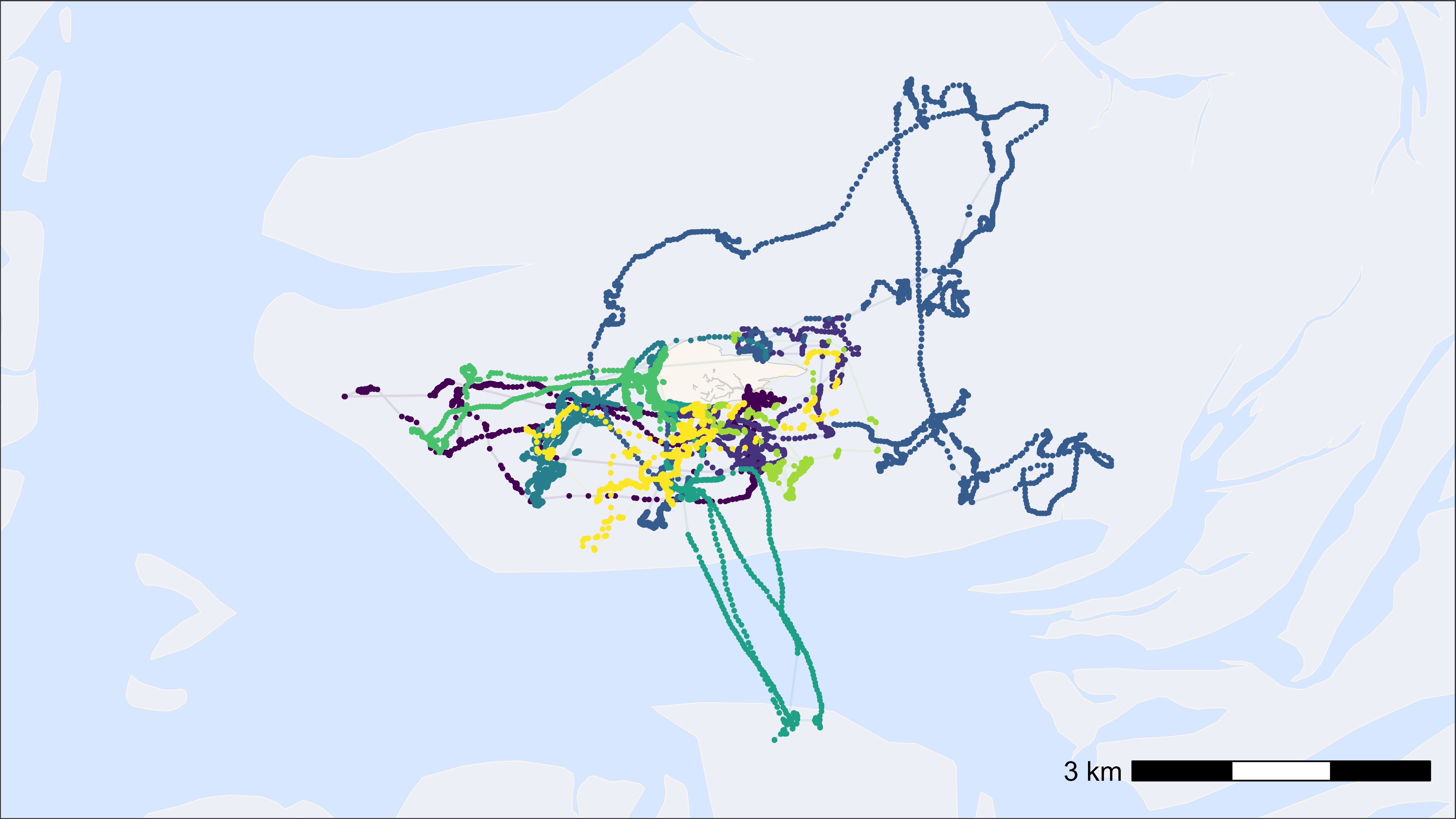
We can also add more information to the label by using the
atl_tag_labs() function. This function can be used to
create labels for the legend that include more information about the
tag, such as metal rings, colour rings or name.
# load example data
data <- data_example
# file path to the metadata
fp <- system.file(
"extdata", "tags_watlas_subset.xlsx", package = "tools4watlas"
)
# load meta data
all_tags <- readxl::read_excel(fp, sheet = "tags_watlas_all") |>
data.table()
# join with metal rings and color rings
all_tags[, tag := as.character(tag)]
data[all_tags, on = "tag", `:=`(
rings = i.rings,
crc = i.crc
)]
# create basemap
bm <- atl_create_bm(data, buffer = 800)
# plot points and tracks with fixed assigned colours (e.g. in animations)
bm +
geom_path(
data = data, aes(x, y, colour = tag),
linewidth = 0.5, alpha = 0.1, show.legend = TRUE
) +
geom_point(
data = data, aes(x, y, colour = tag),
size = 0.5, alpha = 1, show.legend = TRUE
) +
scale_color_manual(
values = atl_tag_cols(data$tag),
labels = atl_tag_labs(data, c("tag", "rings", "crc")),
name = paste("N = ", length(unique(data$tag)))
) +
theme(legend.position = "top")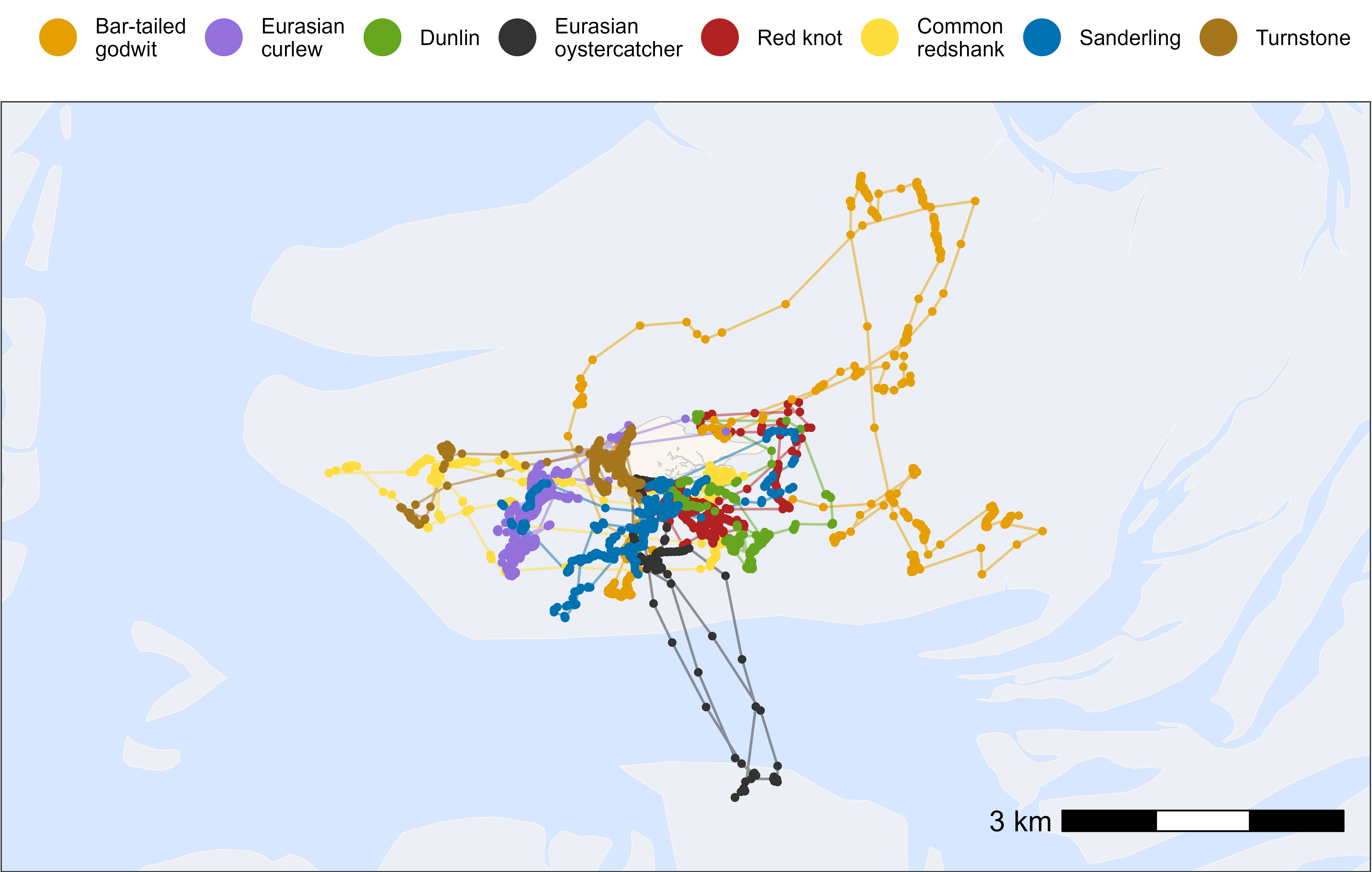
Plot grouped by species
tools4watlas includes specific species colours in the
atl_spec_cols() function and species labels in the atl_spec_labs()
function.
# load example data
data <- data_example
# create basemap
bm <- atl_create_bm(data, buffer = 800)
# plot points and tracks
bm +
geom_path(
data = data, aes(x, y, group = tag, colour = species),
linewidth = 0.5, alpha = 0.5, show.legend = FALSE
) +
geom_point(
data = data, aes(x, y, color = species),
size = 1, alpha = 1, show.legend = TRUE
) +
scale_color_manual(
values = atl_spec_cols(),
labels = atl_spec_labs("multiline"),
name = ""
) +
guides(colour = guide_legend(
nrow = 1, override.aes = list(size = 7, pch = 16, alpha = 1)
)) +
theme(
legend.position = "top",
legend.justification = "center",
legend.key = element_blank(),
legend.background = element_rect(fill = "transparent")
)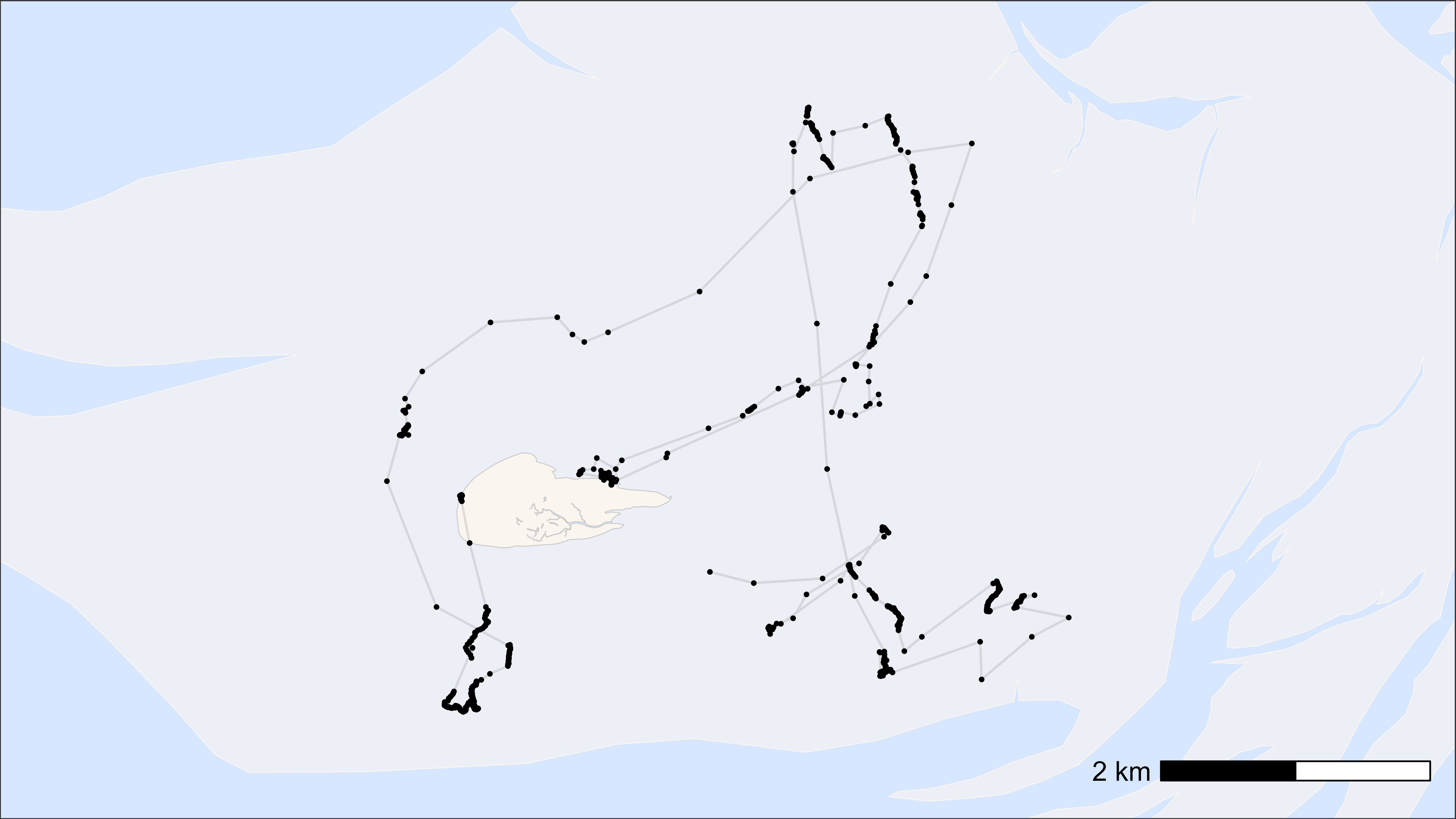
Plot single plots by tag ID
Template for a customizable plot of one tag
# load example data
data <- data_example
# subset bar-tailed godwit
data_subset <- data[species == "bar-tailed godwit"]
# create basemap
bm <- atl_create_bm(data_subset, buffer = 800)
# plot points and tracks
bm +
geom_path(
data = data_subset, aes(x, y),
linewidth = 0.5, alpha = 0.1, show.legend = FALSE
) +
geom_point(
data = data_subset, aes(x, y),
size = 0.5, alpha = 1, show.legend = FALSE
)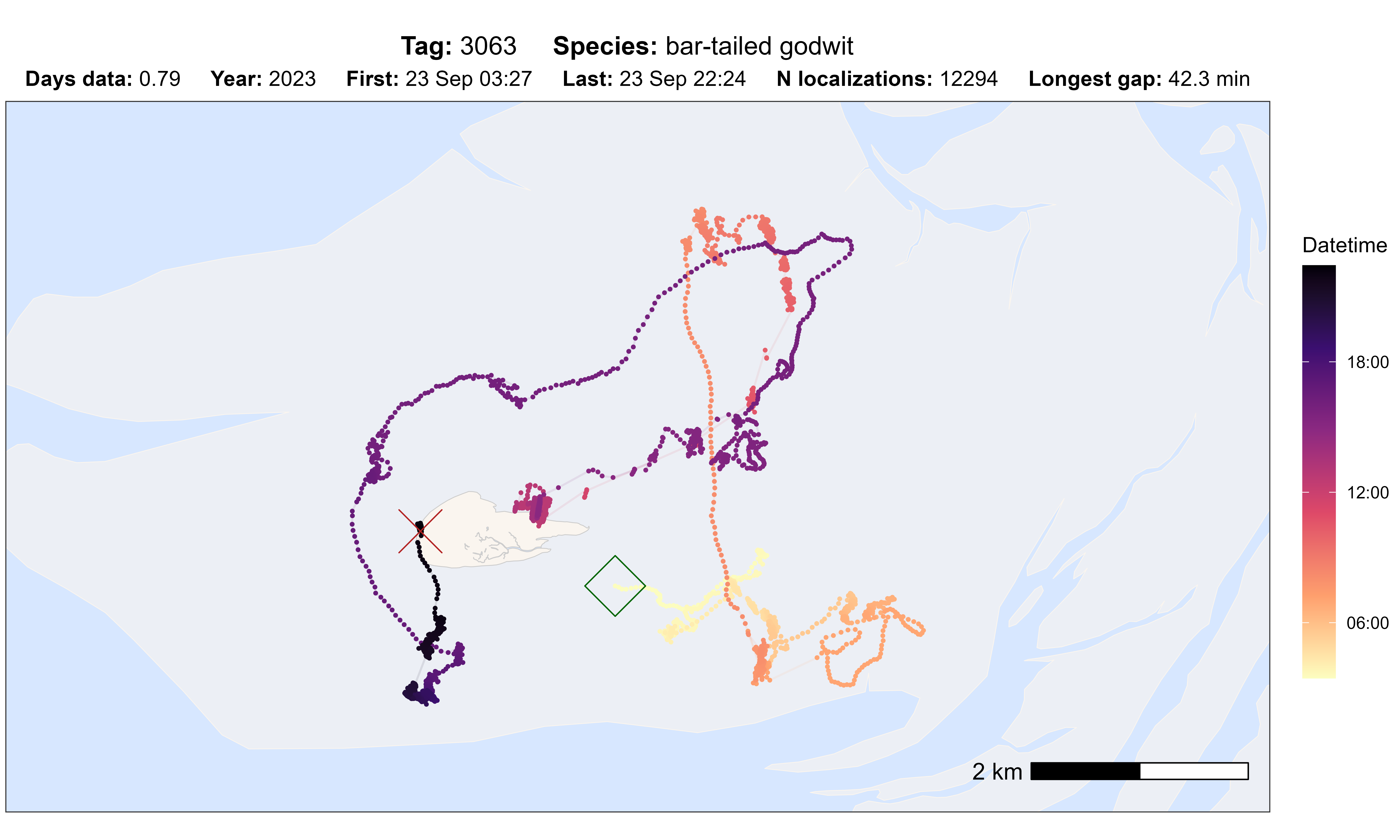
Simple preset plots to check the data
The atl_check_tag() function can be used to quickly
check the data from one tag. It provides five different options for
colouring the track:
-
"datetime": Datetime along the track -
"nbs": Number of receiver (base) stations that contributed to the localization -
"var": Error as maximal variance of varx and vary -
"speed_in": Speed in m/s -
"gap": Gaps coloured by time and as point size
The scale can be coloured with all options from viridis
(default: "A") by specifying the scale_option.
The scale can also be transformed with scale_trans for
example using "log" or "sqrt". First and last
point can be highlighted (highlight_first and
highlight_last) or just a specific number of points from
the beginning or end of the track can be selected (first_n
or last_n).
# path to csv with filtered data
data_path <- system.file(
"extdata", "watlas_data_filtered.csv",
package = "tools4watlas"
)
# load data
data <- fread(data_path, yaml = TRUE)
# subset bar-tailed godwit
data <- data[species == "bar-tailed godwit"]
# plot option datetime
atl_check_tag(
data,
option = "datetime",
highlight_first = TRUE, highlight_last = TRUE
)
# subset with last 1000 positions
atl_check_tag(
data,
last_n = 1000,
option = "datetime",
highlight_first = TRUE, highlight_last = TRUE
)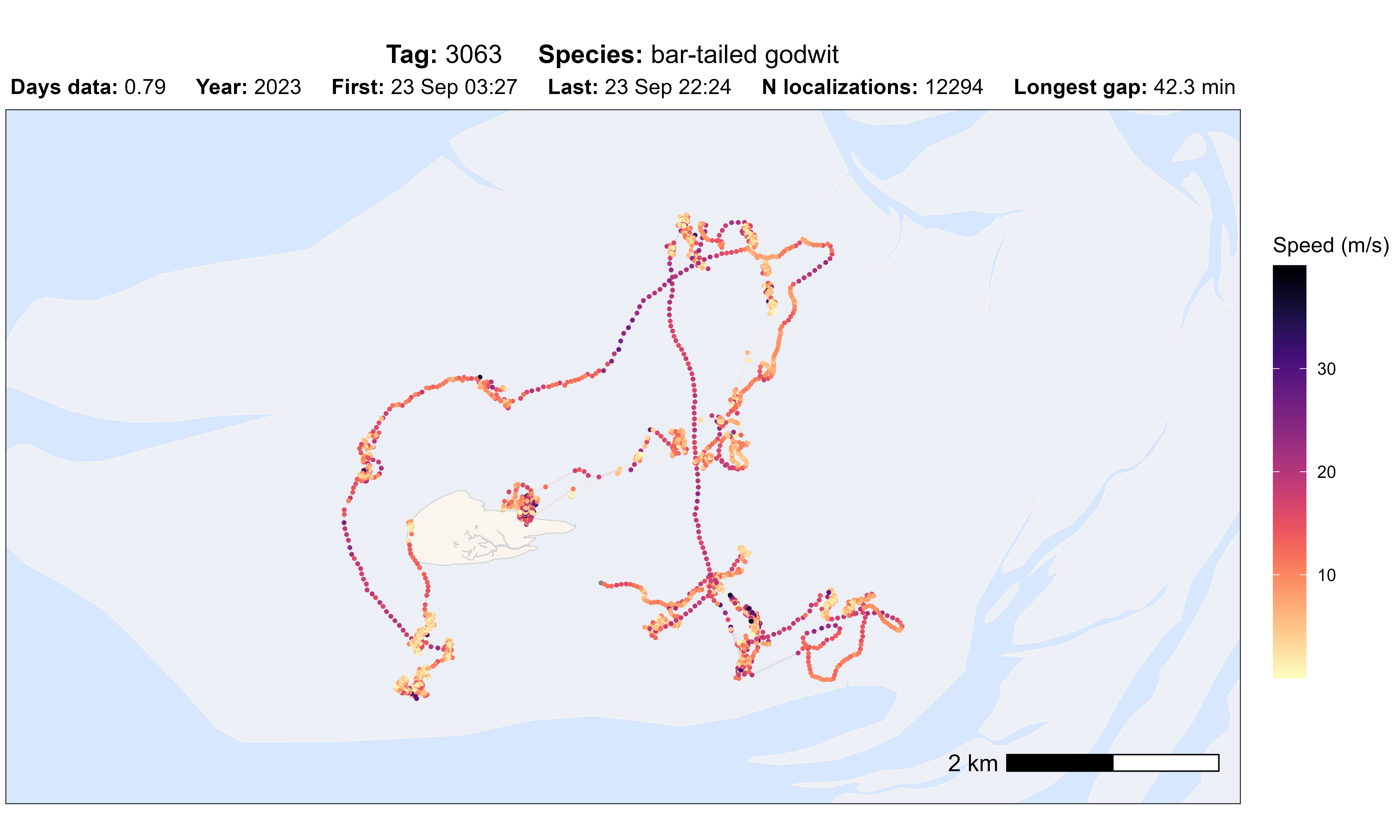
# plot option speed_in
atl_check_tag(data, option = "speed_in")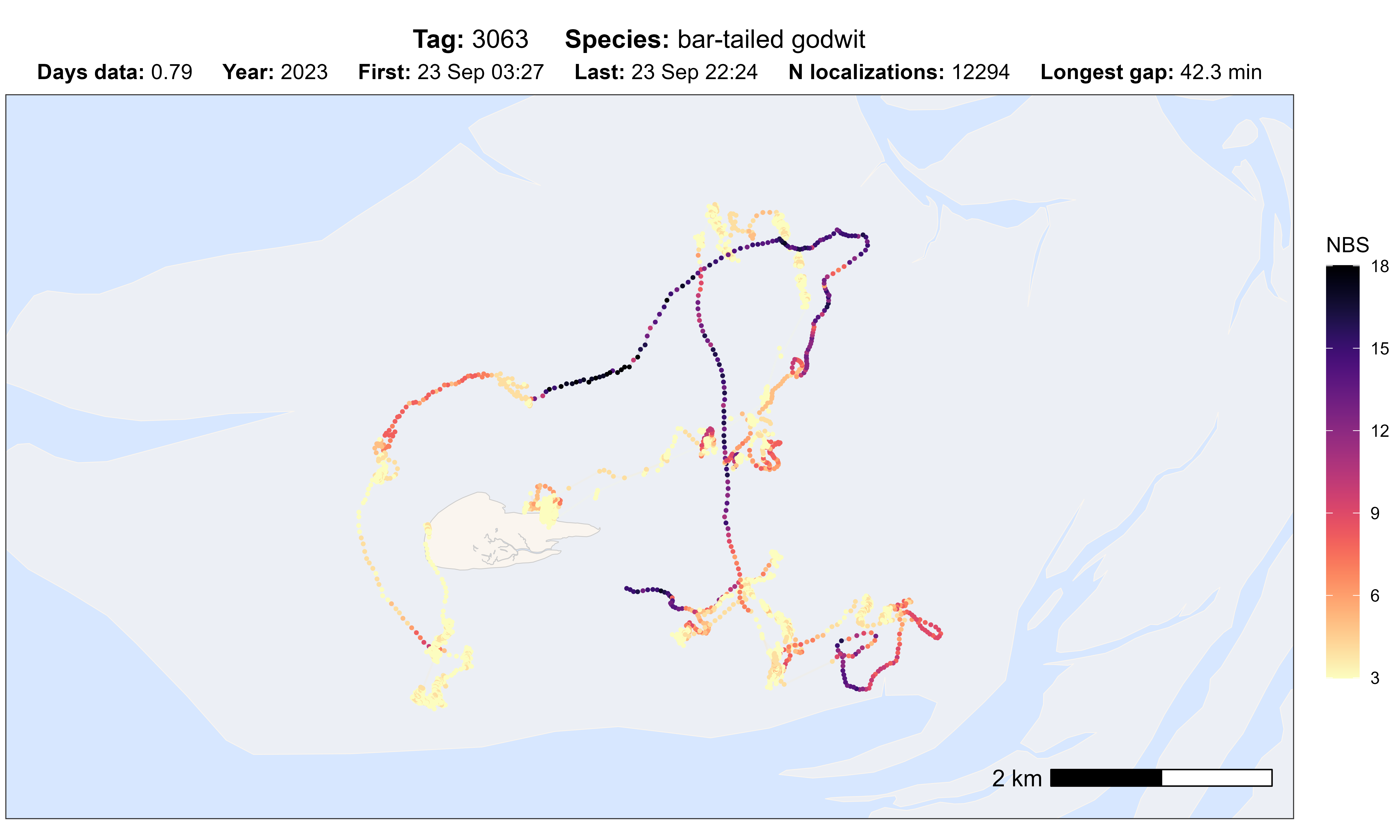
# plot option nbs
atl_check_tag(data, option = "nbs")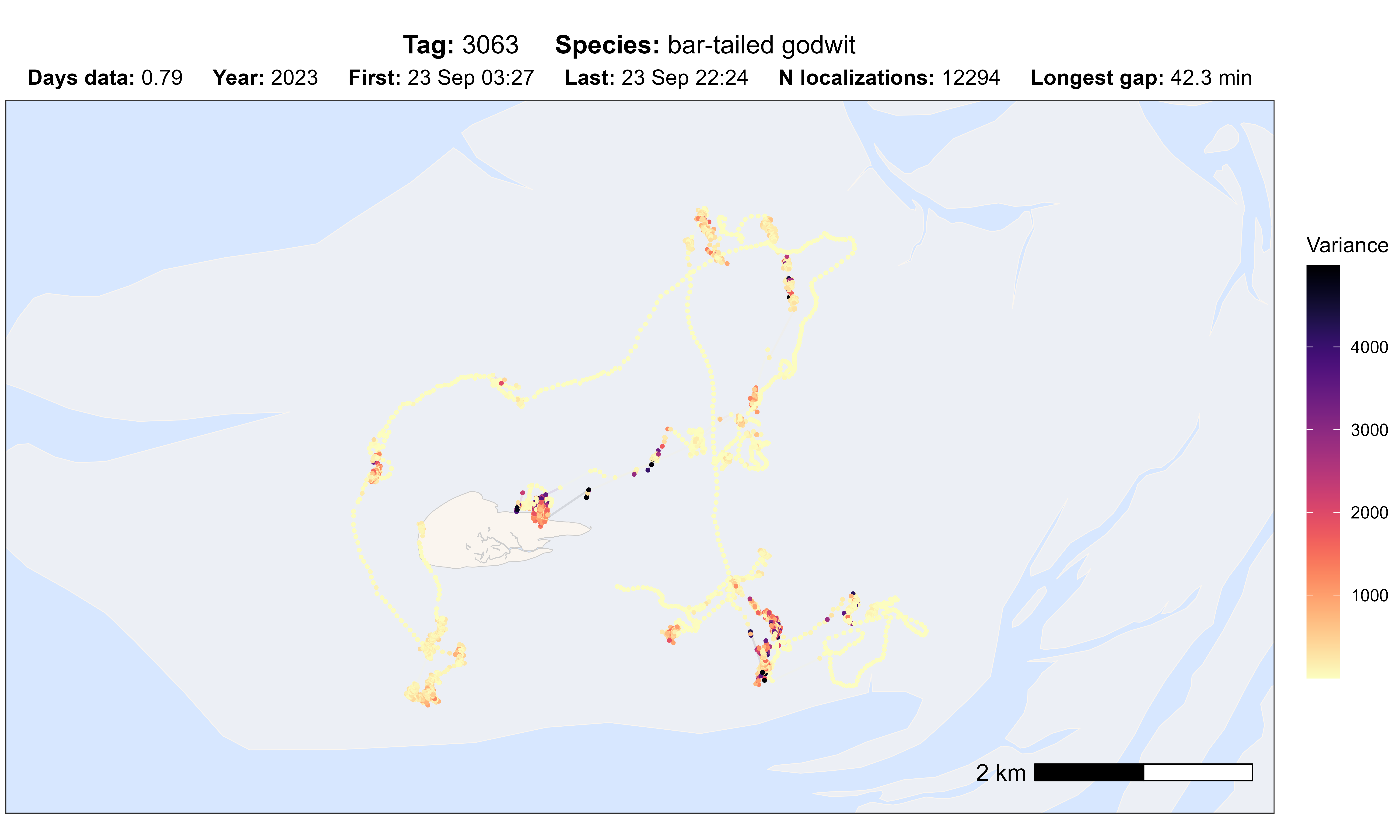
# plot option sd
atl_check_tag(data, option = "var")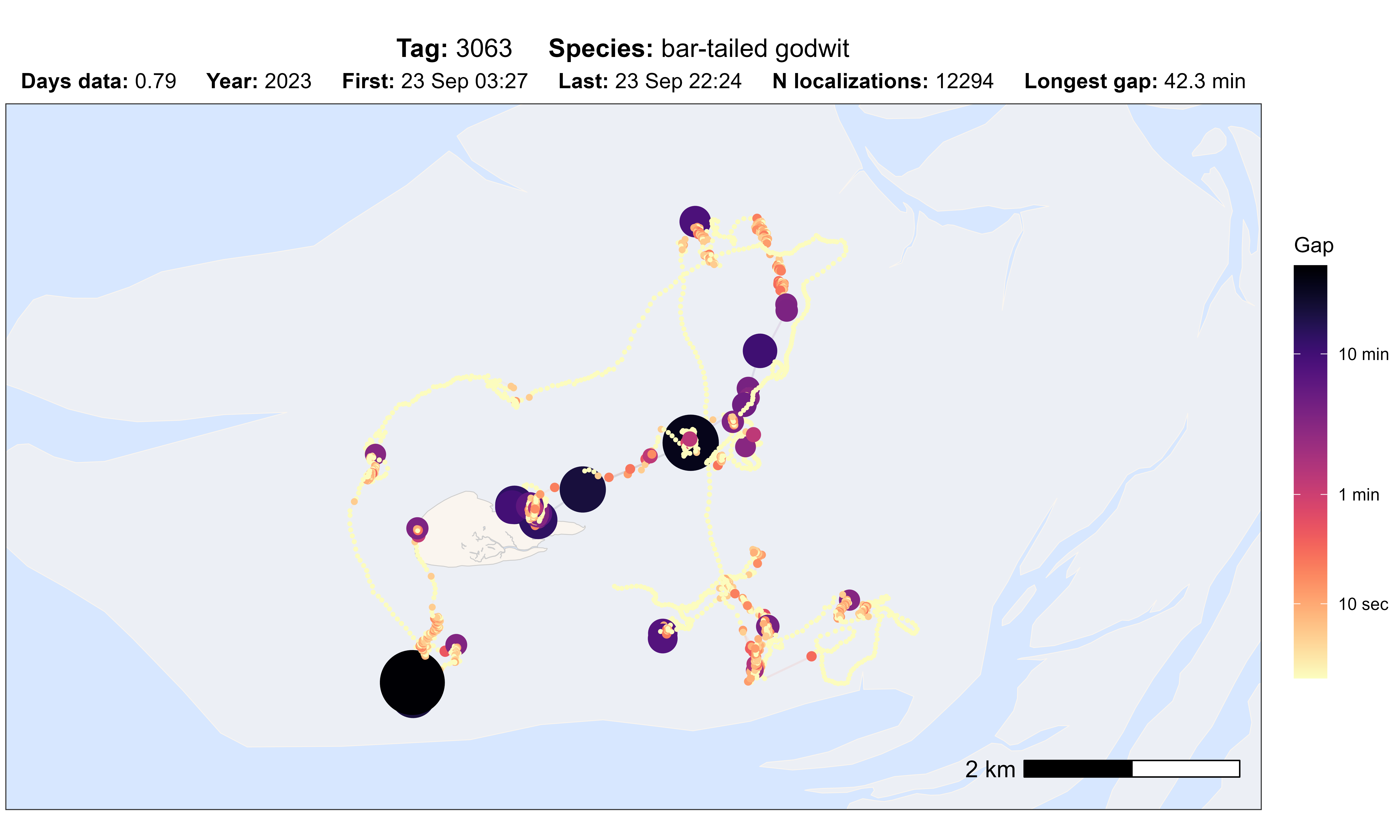
# plot option gap
atl_check_tag(data, option = "gap", scale_trans = "log")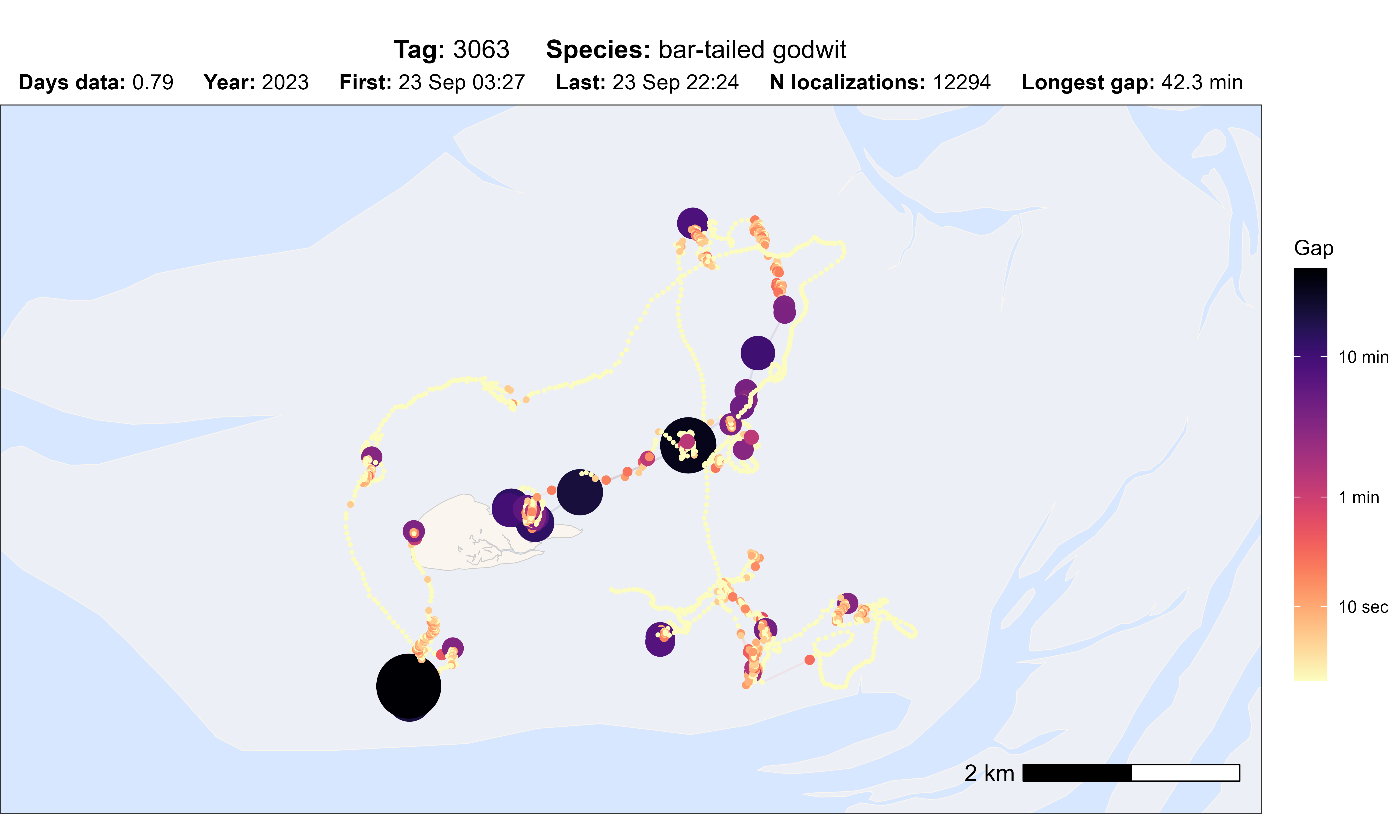
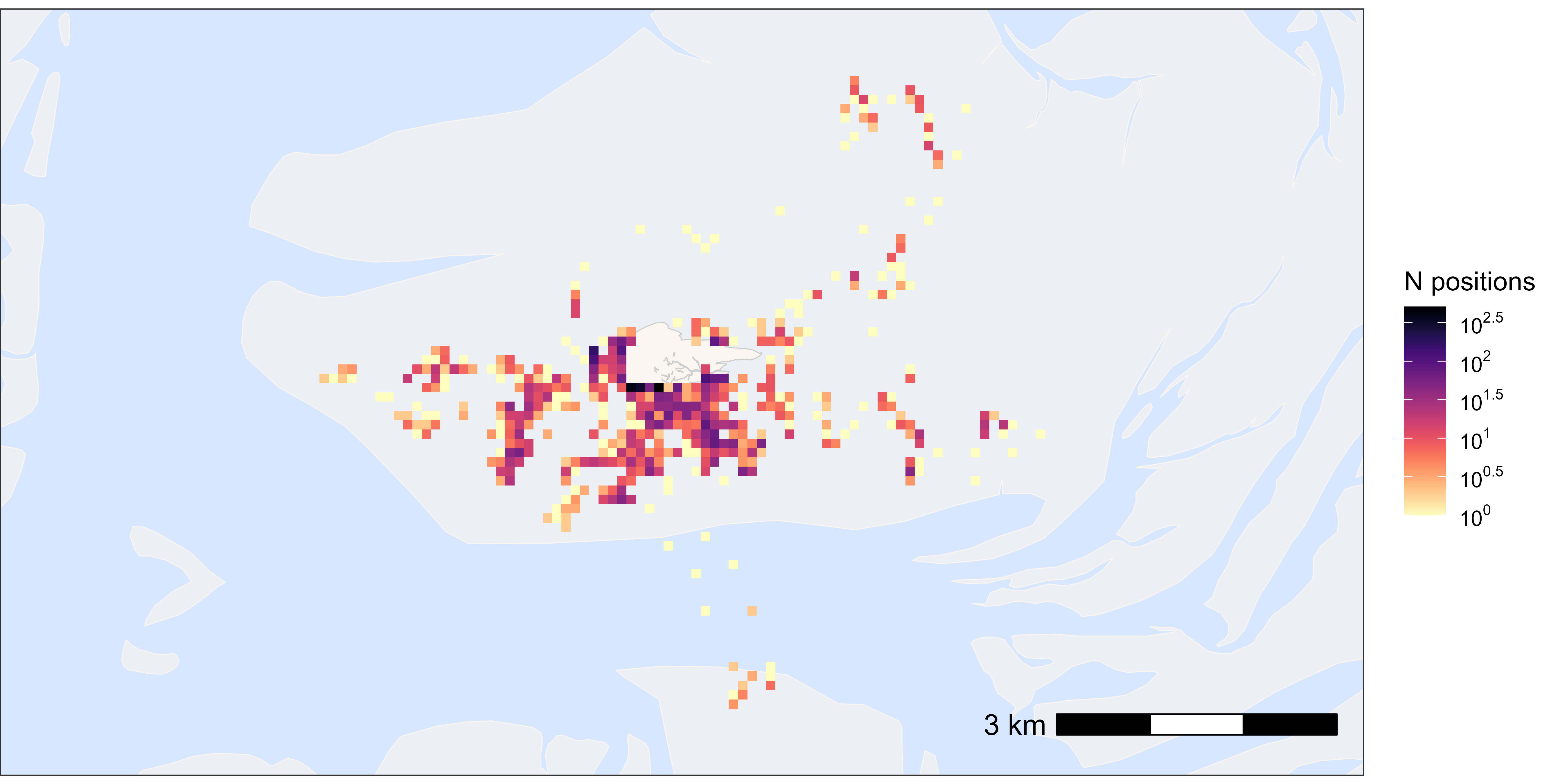
Plot heatmaps
Here we round the data (e.g. to 200 m) and then plot the number of positions per location.
All positions
This is a quick way to get an overview of the data.
# load example data
data <- data_example
# create basemap
bm <- atl_create_bm(data, buffer = 800)
# round data to 1 ha (100x100 meter) grid cells
data[, c("x_round", "y_round") := list(
plyr::round_any(x, 100),
plyr::round_any(y, 100)
)]
# N by location
data_subset <- data[, .N, by = c("x_round", "y_round")]
# plot heatmap
bm +
geom_tile(
data = data_subset, aes(x_round, y_round, fill = N),
linewidth = 0.1, show.legend = TRUE
) +
scale_fill_viridis(
option = "A", discrete = FALSE, trans = "log10", name = "N positions",
breaks = trans_breaks("log10", function(x) 10^x),
labels = trans_format("log10", math_format(10^.x)),
direction = -1
)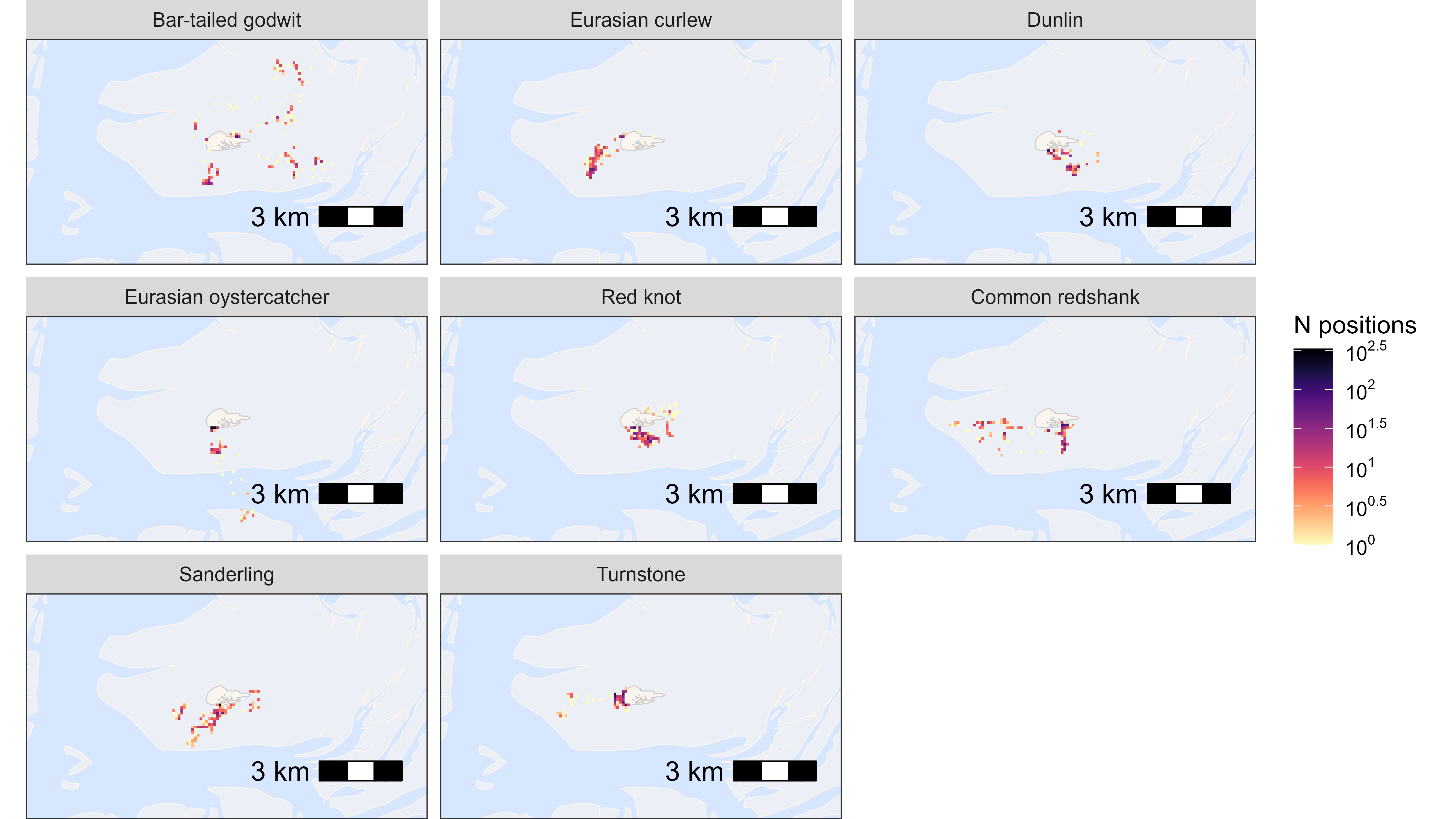
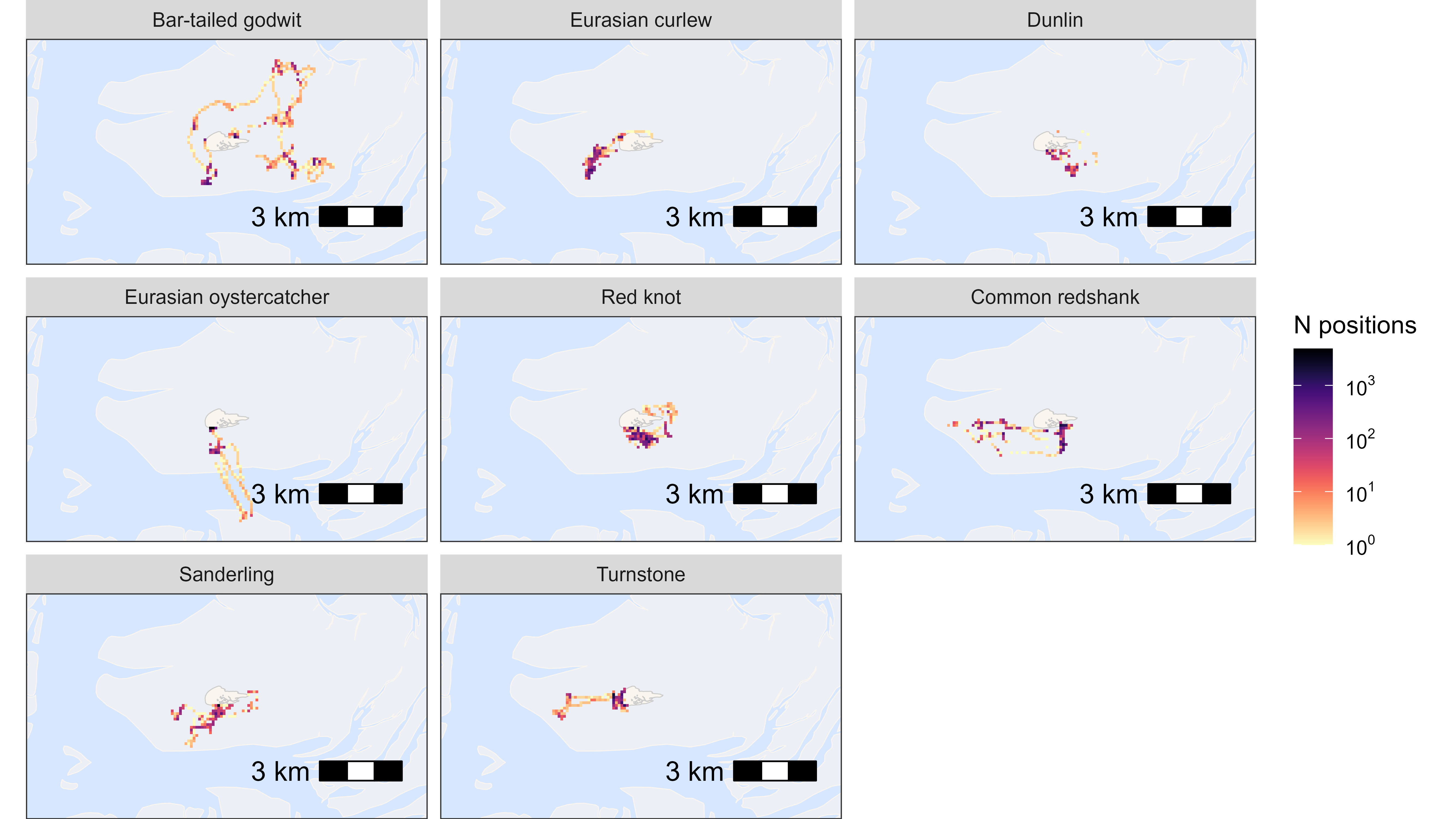
Positions by species
We group the data by species (or whatever group we desire) and then make a plot for each of those groups.
# N by location and species
data_subset <- data[, .N, by = c("x_round", "y_round", "species")]
# plot heatmap by species
bm +
geom_tile(
data = data_subset, aes(x_round, y_round, fill = N),
linewidth = 0.1, show.legend = TRUE
) +
scale_fill_viridis(
option = "A", discrete = FALSE, trans = "log10", name = "N positions",
breaks = trans_breaks("log10", function(x) 10^x),
labels = trans_format("log10", math_format(10^.x)),
direction = -1
) +
facet_wrap(~species, labeller = as_labeller(atl_spec_labs("singleline")))